Recently, non-homogenous tissue based methods have been developed for proteomic and lipidomic analysis, and they appear to be reliable for tumor classification for digestive, brain, lymphomatous, and lung cancers. Among these tissue-based methods, MALDI-Tubuloside-A imaging is now used by several teams for clinical research. However, the MALDI-imaging approach remains complex because it requires frozen tissue slice analysis results to co-register MALDI spectra imaging and morphology imaging. For human liver metastasis samples, this method allowed tumor classification into six common cancer types with a sensitivity varying from 54% to 88%, and a specificity varying from 90% to 98% depending on the malignant class. To simplify the process, Lee and coauthors proposed performing MALDI-ToF MS for lipidomics analysis of preselected frozen section slices containing at least 70% malignant cells. The resulting spectra were used to generate a modelthat accurately classified normal lung tissues, lung tumor tissues, and primary NSCLC. Primary NSCLC was accurately discriminated from other types of lung tumors, and the three subclasses, adenocarcinoma, squamous-cell and large-cell carcinoma, were correctly discriminated and classified with a sensitivity and a specificity of 84% and 77%, respectively for adenocarcinoma versus squamous cell carcinoma. The authors recorded no misclassified sample when comparing Primary NSCLC and other types of lung tumors, whereas in the present study, we found both false negatives and false positives when we compared Primary lung cancer versus Metastasis subclasses. The difference in our study sample size, with greater numbers of tumoral and non-tumoral samplescompared to the above-mentioned study, could explain differences in diagnostic performance results. In addition, good diagnostic performance from other studies was achieved by applying MALDI-imaging on chosen regions that contained high tumor cellularitybased on the histology of sections stained with hematoxylin and eosin. Here, we used no pre-selection of tissue samples and obtained good results. We targeted tumoral pieces larger than 1 cm which represent the most frequent surgical indications. It is plausible that the size of the tumor have favorably influenced our results since the risk of having sampled a bad territory is reduced with large Epimedoside-A tumors as compared to millimeter tumors. Mass spectrometry imaging strategies offer the 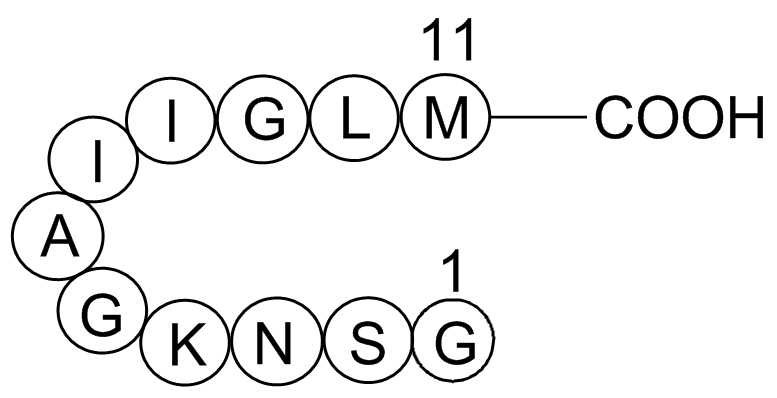 advantage of conserving tissue but require sufficient surface area of tissue sections to obtain valuable information. In addition, MS imaging methods require trained experts, heavy analysis software and highthroughput signal acquisition instrumentation. Like these abovementioned methods, our strategy did not require any purification or standardization of the tissue cell content. Our crushed sample MS analysis was rapid, reproducible and very easy to perform. The non-conservative aspect of our approach was in part counterbalanced by the very low tissue sample sizeable to give valid spectra. Finally, using a simplified and non-image-guided method and larger cohort of patients, we obtained diagnostic performances similar to those obtained with MALDI-imaging methods or purified cell line methods. This surprising result could be due to more the complete information contained in complete unpurified tissue sample and to our modest objective, which was not to identify the exact nature of the tumor but to classify the sample into either the Cancer or non-tumor class.
advantage of conserving tissue but require sufficient surface area of tissue sections to obtain valuable information. In addition, MS imaging methods require trained experts, heavy analysis software and highthroughput signal acquisition instrumentation. Like these abovementioned methods, our strategy did not require any purification or standardization of the tissue cell content. Our crushed sample MS analysis was rapid, reproducible and very easy to perform. The non-conservative aspect of our approach was in part counterbalanced by the very low tissue sample sizeable to give valid spectra. Finally, using a simplified and non-image-guided method and larger cohort of patients, we obtained diagnostic performances similar to those obtained with MALDI-imaging methods or purified cell line methods. This surprising result could be due to more the complete information contained in complete unpurified tissue sample and to our modest objective, which was not to identify the exact nature of the tumor but to classify the sample into either the Cancer or non-tumor class.
Obtained using spectral patterns from a homogenous population of cell suspensions
Leave a reply