To remove the variant surface protein from the membrane, required for parasite differentiation into the procyclic insect form. Gp63-like molecules have been observed on the cell surface of symbiont-harboring trypanosomatids. Importantly, the symbiont containing A. deanei displays a higher amount of leishmanolysin-like molecules at the surface compared to the aposymbiotic strain, which are unable to colonize insects. As anti-gp63 antibodies decrease protozoan-insect interactions, our results reinforce 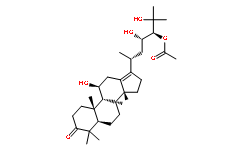 the idea that the presence of such interactions caused the expansion of this gene family in endosymbiont-bearing organisms. This class of cysteine peptidase is represented by cruzain or cruzipain, major lysosomal proteinases of T. cruzi expressed by parasites found in insect and vertebrate hosts, and encoded by a large gene family. In T. cruzi, these enzymes have important roles in various aspects of the host/parasite relationship and in intracellular digestion as a nutrient source. Conversely, the low copy number of this class of lysosomal peptidase in symbiont-containing trypanosomatids seems to be related to their low nutritional requirements. Amastins constitute a third large gene family in the A. deanei and S. culicis genomes that encodes surface proteins. Initially described in T. cruzi, amastin genes have also been identified in various Leishmania species, in A. deanei and in another related insect parasite, Leptomonas seymouri. In Leishmania, amastins constitute the largest gene family with gene expression that is regulated during the parasite life cycle. As amastin has no sequence similarity to any other known protein, its function remains unknown. In this work, we identified 31 genes with sequences belonging to all four Orbifloxacin sub-families of amastins in the genome of A. deanei and 14 copies of amastin genes in S. culicis. Similar to Leishmania, members of all four amastin subfamilies were identified in symbiont-containing species. The putative proteome of symbiont-bearing trypanosomatids revealed that these microorganisms exhibit unique features when compared to other protozoa of the same family and that they are most closely related to Leishmania species. Most relevant are the differences in the genes related to cytoskeleton, paraflagellar and kinetoplast structures, along with a unique pattern of peptidase gene organization that may be related to the presence of the symbiont and of the monoxenic life style. The symbiotic bacteria of A. deanei and S. culicis are phylogenetically related with a common ancestor, most likely a b-proteobacteria of the Alcaligenaceae family. The genomic content of these symbionts is highly reduced, indicating gene loss and/or transfer to the host cell nucleus. In addition, we confirmed that both bacteria contain genes that encode enzymes that complement several Folinic acid calcium salt pentahydrate metabolic routes of the host trypanosomatids, supporting the fitness of the symbiotic relationship. Aerobic life uses the oxidizing power of O2 for various fundamental biochemical processes including energy generation in form of ATP synthesis realised by a controlled stepwise reduction of molecular oxygen. However, oxygen consumption has severe side effects, since, inevitably, reactive oxygen species are produced as by-products. If not countered or buffered, these ROS attack and disturb biological molecules and cellular structures leading to a state of impaired physiology also known as oxidative stress.
the idea that the presence of such interactions caused the expansion of this gene family in endosymbiont-bearing organisms. This class of cysteine peptidase is represented by cruzain or cruzipain, major lysosomal proteinases of T. cruzi expressed by parasites found in insect and vertebrate hosts, and encoded by a large gene family. In T. cruzi, these enzymes have important roles in various aspects of the host/parasite relationship and in intracellular digestion as a nutrient source. Conversely, the low copy number of this class of lysosomal peptidase in symbiont-containing trypanosomatids seems to be related to their low nutritional requirements. Amastins constitute a third large gene family in the A. deanei and S. culicis genomes that encodes surface proteins. Initially described in T. cruzi, amastin genes have also been identified in various Leishmania species, in A. deanei and in another related insect parasite, Leptomonas seymouri. In Leishmania, amastins constitute the largest gene family with gene expression that is regulated during the parasite life cycle. As amastin has no sequence similarity to any other known protein, its function remains unknown. In this work, we identified 31 genes with sequences belonging to all four Orbifloxacin sub-families of amastins in the genome of A. deanei and 14 copies of amastin genes in S. culicis. Similar to Leishmania, members of all four amastin subfamilies were identified in symbiont-containing species. The putative proteome of symbiont-bearing trypanosomatids revealed that these microorganisms exhibit unique features when compared to other protozoa of the same family and that they are most closely related to Leishmania species. Most relevant are the differences in the genes related to cytoskeleton, paraflagellar and kinetoplast structures, along with a unique pattern of peptidase gene organization that may be related to the presence of the symbiont and of the monoxenic life style. The symbiotic bacteria of A. deanei and S. culicis are phylogenetically related with a common ancestor, most likely a b-proteobacteria of the Alcaligenaceae family. The genomic content of these symbionts is highly reduced, indicating gene loss and/or transfer to the host cell nucleus. In addition, we confirmed that both bacteria contain genes that encode enzymes that complement several Folinic acid calcium salt pentahydrate metabolic routes of the host trypanosomatids, supporting the fitness of the symbiotic relationship. Aerobic life uses the oxidizing power of O2 for various fundamental biochemical processes including energy generation in form of ATP synthesis realised by a controlled stepwise reduction of molecular oxygen. However, oxygen consumption has severe side effects, since, inevitably, reactive oxygen species are produced as by-products. If not countered or buffered, these ROS attack and disturb biological molecules and cellular structures leading to a state of impaired physiology also known as oxidative stress.
Monthly Archives: June 2019
The Hif1a transcription factor suggesting that Hif1a can directly regulate their transcription
Because Hif1a functions as a transcriptional activator, it likely exerts its protective effects through increased expression of genes involved in DNA repair or cell survival. Previous studies indicate that hypoxia can alter expression of components of the mismatch repair pathway; however, altered expression of these proteins would not account for radioprotection observed after activation of Hif1a by DMOG. However, we identified 3 new targets for Hif1a – the CHD4 helicase, the MTA3 regulatory protein and the Suv39h1 methyltransferase. All 3 were increased at both the mRNA and protein level by DMOG and the hypoxia mimetic agent CoCl2. Previous work has shown that MTA1, a close family member related to MTA3, is also transcriptionally upregulated by Hif1a during hypoxia, implying that the MTA family of coactivators are a common target for the Hif1a transcription factor. Both CHD4 and MTA3 are components of the NuRD complex, a histone deacetylase complex which is implicated in DNA repair. Inactivation of CHD4 leads to increased sensitivity to IRinduced DNA damage, indicating a key role in the repair of DSBs. Taken together, this would suggest that increased levels of NuRD may protect cells from radiation. However, although Hif1a was important for DMOG to protect cells from IR, loss of either CHD4 or MTA3 expression did not alter sensitivity to IR in our cell system. We interpret this to mean that, although DMOG can stabilize Hif1a and increase levels of the NuRD complex, the accumulation of NuRD does not significantly impact the radiosensitivity of the cells. The accumulation of the NuRD deacetylase complex may therefore play an alternate role in the hypoxia response, such as transcriptional repression of genes during low oxygen tension. An alternate explanation for how DMOG may protect cells from radiation damage can be proposed based on the previous observation that Hif1a can alter expression of genes involved in the regulation of histone methylation. Hif1a increases expression of histone demethylases, including KDM4A and KDM4B, which function to remove methyl groups from methylated histones on the chromatin. The increased expression of KDMs during hypoxia is associated with a decrease in histone methylation, including a reduction on H3K9me3 levels. Previous work has shown that a decrease in H3K9me3 levels is associated with an increase in radiosensitivity. Hif1a -dependent increases in KDM expression would therefore be predicted to increase, XAV939 rather than decrease, radiosensitivity. However, we found that, unlike hypoxia, DMOG increased rather than decreased H3K9me3 levels. This result is explained by the fact that KDMs and Hif1a-prolylhydroxylases share a common catalytic mechanism, so that both classes of enzyme are inhibited by DMOG. DMOG therefore inhibits the Hif1a-prolylhydroxylase, leading to accumulation of transcriptionally active Hif1a and upregulation of KDMs. However, because DMOG can also directly  inhibit KDMs, this essentially negates the increased expression of KDMs VE-821 moa mediated by Hif1a. Overall, DMOG will increase histone methylation through inhibition of KDMs.
inhibit KDMs, this essentially negates the increased expression of KDMs VE-821 moa mediated by Hif1a. Overall, DMOG will increase histone methylation through inhibition of KDMs.
There are clear differences among viruses on their requir inhibition of any of those regulators causes a halt in internalization
It remains thus possible that inhibition of efficient assembly of these signaling proteins or inhibition of activation of any of these factors could be contributing to the block in uptake. Cholesterol-rich membrane domains have been shown to mediate entry of enveloped flaviviruses such as dengue virus and Japanese encephalitis virus. In addition, filipin was shown to block HIV gag protein entry and translocation to MVBs in a clathrin-independent pathway. We found here that choleTulathromycin B sterol domains were also needed for the biogenesis of cytoplasmic EV1-induced multivesicular bodies. Ketoconazole treatment, which caused accumulation of lanosterol and lowered the amount of cholesterol in cells, caused integrin depletion out from detergent-resistant domains and had a drastic effect on internalization. The endosomal structures that were observed in the cytoplasm were early endosomal type tubulovesicular structures, suggesting that sufficient content of cholesterol in the membranes is needed for the formation of multivesicularity. In line with these results, the deletion of yeast OSH genes encoding oxysterol-binding proteins alters intracellular sterol distribution and causes vacuolar fragmentation. On the other hand, overexpression of one sterol lipid carrier ORP1L was shown to cause enlarged multivesicular bodies with higher amount of internal membranes altogether suggesting that sterol abundance and distribution is important for the biogenesis and function of endosomes. We showed further that cholesterol-rich domains are cointernalized and exist in the formed a2-MVBs. This was proven by cholesterol labeling with filipin and by the classical cold Triton X-100 treatment, which had no solubilizing effect on the a2MVBs whereas it had a more pronounced effect in the control endosomal structures labeled and induced by clustered aV integrin. This was expected as we had recently shown that cholesterol binding caveolin-1 rich domains are also accumulating in a2-MVBs. We also knew from our previous studies that clustered aV integrin is internalized via clathrin coated pits and Folinic acid calcium salt pentahydrate colocalizing with EEA1 and transferrin unlike EV1. Although it has been shown that different endosomes contain lipid raft components, our results suggest that a2-MVBs, originated from the cholesterol-rich plasma membrane domains contain a higher amount of detergent-insoluble lipids than the antibody clustered aV integrin structures studied here. Experiments with filipin and nystatin clearly showed that EV1 inside a2-MVBs was sensitive to these drugs. Administration of these drugs halted virus infection at the a2-MVBs. While drug addition during the active replication period was not much affected, administration during the early time points totally prevented infection. The later replication phase has been shown to be sensitive for cholesterol perturbation for hepatitis C virus, West Nile virus and Japanese encephalitis virus. In the case of  West Nile virus, virus causes redistribution of cholesterol from the plasma membrane and recruits cholesterol to the dsRNA-positive replication sites. Hepatitis C virus RNA was shown to accumulate in detergent-insoluble vesicles and to colocalize with caveolin-2. Also flaviviral non-structural proteins were shown to associate with detergent-resistant membranes. In contrast, labeling of dsRNA during EV1 replication showed that indeed, dsRNA-positive replication sites did not accumulate cholesterol, further suggesting that the replication step of EV1 is not particularly dependent on membranes enriched in cholesterol.
West Nile virus, virus causes redistribution of cholesterol from the plasma membrane and recruits cholesterol to the dsRNA-positive replication sites. Hepatitis C virus RNA was shown to accumulate in detergent-insoluble vesicles and to colocalize with caveolin-2. Also flaviviral non-structural proteins were shown to associate with detergent-resistant membranes. In contrast, labeling of dsRNA during EV1 replication showed that indeed, dsRNA-positive replication sites did not accumulate cholesterol, further suggesting that the replication step of EV1 is not particularly dependent on membranes enriched in cholesterol.
Reflected by variation in MEICS-score and expression of the pro-inflammatory cytokines IL-1a and IL-1b
TNBS-ethanol administration is characterized by Th1-driven inflammation, and has primarily been regarded as a model for CD. The clinical features include weight loss and bloody diarrhea, while morphologically the model is characterized by mucosal, subMepiroxol mucosal and transmural inflammation. Consequently, the model has been used to investigate the role and 3,4,5-Trimethoxyphenylacetic acid mechanistic action of drugs including 5-aminosalisylic acid, steroids and  anti-tumor necrosis factor in IBD. The administration and doses of TNBS differ significantly between studies; the methodology is inconsistent and no standardized protocol exists. Both TNBS-colitis and IBD include disturbances in basic physiological processes like immune activation, metabolism and mucosal repair. Microbial regulation of TLRs and induction of an inflammatory response is accompanied by regulation of proand anti-inflammatory cytokines and the shaping of the intestinal immune response. Down-regulation of the peroxisome proliferator-activated receptor gamma involved in the regulation of fatty acid metabolism and inflammation, has been demonstrated in IBD and is associated with maintenance of defensin expression. The cellular Prion protein is expressed in brain and various extra cerebral tissues including neuroendocrine cells and lymphoid tissue of the gut. PrPc can have an anti-inflammatory effect in the colon. We report the development and characterization of TNBScolitis in rats using colonoscopy and temporal gene expression profiling. The aim was to standardize TNBS administration to achieve a moderate inflammation. Achieving a moderate colitis allowed for the study of epithelial alterations upon mucosal damage, while temporal gene expression profiling identified longitudinal transcriptomic changes that were associated with these abnormalities. We further aimed to compare the genomewide changes in TNBS-colitis to a human transcriptome to determine whether TNBS at this dose was an appropriate model for IBD. TNBS-colitis is widely used as a model for IBD. However, its resemblance to human disease has not been thoroughly explored. TNBS-colitis in rats was originally described as a model for induction of long lasting inflammation and ulceration of the rat colon, and the reproducibility was emphasized. The reproducibility and duration/chronicity of inflammation has, however, been subject to debate. In the current study, we standardized the protocol for induction of moderate TNBS-colitis in Sprague Dawley rats and evaluated temporal changes in gene expression profiles and biological pathways after the induction of colitis. Importantly, we quantitatively assessed concordance of the TNBS-colitis and IBD transcriptomes. Doses and concentrations of TNBS used in previous studies differ markedly and no standardized protocol has been generally developed. We therefore aimed to optimize the method to achieve a reproducible moderate colitis. High concentrations of TNBS, administered in small volumes induced a localized severe inflammation resulting in deep colonic ulcerations, coprostasis, stenoses and ileus. Milder inflammation and more wide spread inflammation was achieved with lower TNBS concentrations in a larger total volume. A TNBS concentration of 30 mg/ml in a total volume of 0.6 ml resulted in a moderate inflammation involving most of the left colon. However, despite the use of a standardized protocol, a slight inter-individual dissimilarity in the extension and degree of inflammation was seen.
anti-tumor necrosis factor in IBD. The administration and doses of TNBS differ significantly between studies; the methodology is inconsistent and no standardized protocol exists. Both TNBS-colitis and IBD include disturbances in basic physiological processes like immune activation, metabolism and mucosal repair. Microbial regulation of TLRs and induction of an inflammatory response is accompanied by regulation of proand anti-inflammatory cytokines and the shaping of the intestinal immune response. Down-regulation of the peroxisome proliferator-activated receptor gamma involved in the regulation of fatty acid metabolism and inflammation, has been demonstrated in IBD and is associated with maintenance of defensin expression. The cellular Prion protein is expressed in brain and various extra cerebral tissues including neuroendocrine cells and lymphoid tissue of the gut. PrPc can have an anti-inflammatory effect in the colon. We report the development and characterization of TNBScolitis in rats using colonoscopy and temporal gene expression profiling. The aim was to standardize TNBS administration to achieve a moderate inflammation. Achieving a moderate colitis allowed for the study of epithelial alterations upon mucosal damage, while temporal gene expression profiling identified longitudinal transcriptomic changes that were associated with these abnormalities. We further aimed to compare the genomewide changes in TNBS-colitis to a human transcriptome to determine whether TNBS at this dose was an appropriate model for IBD. TNBS-colitis is widely used as a model for IBD. However, its resemblance to human disease has not been thoroughly explored. TNBS-colitis in rats was originally described as a model for induction of long lasting inflammation and ulceration of the rat colon, and the reproducibility was emphasized. The reproducibility and duration/chronicity of inflammation has, however, been subject to debate. In the current study, we standardized the protocol for induction of moderate TNBS-colitis in Sprague Dawley rats and evaluated temporal changes in gene expression profiles and biological pathways after the induction of colitis. Importantly, we quantitatively assessed concordance of the TNBS-colitis and IBD transcriptomes. Doses and concentrations of TNBS used in previous studies differ markedly and no standardized protocol has been generally developed. We therefore aimed to optimize the method to achieve a reproducible moderate colitis. High concentrations of TNBS, administered in small volumes induced a localized severe inflammation resulting in deep colonic ulcerations, coprostasis, stenoses and ileus. Milder inflammation and more wide spread inflammation was achieved with lower TNBS concentrations in a larger total volume. A TNBS concentration of 30 mg/ml in a total volume of 0.6 ml resulted in a moderate inflammation involving most of the left colon. However, despite the use of a standardized protocol, a slight inter-individual dissimilarity in the extension and degree of inflammation was seen.
RETNLBdeficiency increases intestinal permeability and RETNLB seems immune response
Partly be explained by tolerance development and polarization differences in TLR-receptor distribution. PRNP mRNA was significantly up-regulated in TNBScolitis. This was verified by ISH which showed expression in submucosal immune cells and some expression in epithelial cells in animals not yet exposed to TNBS, but expression was more pronounced after colitis induction both in the submucosal immune cells and within the epithelium. PrPc is expressed in several tissues, Mepiroxol including neural tissue, gut-associated lymphoid tissue, enteroendocrine cells and enterocytes. Its function is not well known. Expression of PrPc in the host is thought to be necessary for the propagation and transmission of prion infection. In addition, both pro- and anti-inflammatory roles have been suggested. Reduced inflammatory infiltration has been seen in PrPc-deficient mice upon stimulation with TLR2 and TLR4 ligands. In contrast, PrPc over-expressing mice are resistant to colitis induction with DSS while PrPc deficient mice had significantly higher levels of inflammatory cytokines compared to DSS-treated wild type mice. Increased intestinal barrier permeability has been demonstrated in PrPc-deficient mice, Folinic acid calcium salt pentahydrate further supporting a protecting role for PrPc in colitis and IBD. PPARc is one of three identified isoforms of peroxisome proliferator-activated receptors and is expressed in several tissues; expression is high in the colonic epithelial cells. It is thought to have an important role in modulation of inflammation. PPARc suppresses inflammatory cytokines like TNFa, interleukin6 and IL-1b and maintains defensin production. Gene expression analysis revealed down-regulation of PPARc after colitis induction. In endoscopic biopsies evaluated by ISH, a high basal expression of PPARc in non-inflamed colonic epithelium, while a considerable down-regulation in inflamed colon epithelium was identified. This was accompanied by downregulation of FABP1. FABP1 transcription, in the microarray was confirmed by PCR. The intracellular level of FABP1 positively regulates PPARc and the two proteins also have a direct proteinprotein interaction. Regulation of FABPs is seen in IBD, PPARc is down-regulated in UC and gene polymorphisms of PPARc have been implicated in the pathogenesis of CD. PPARc-agonists have been shown to ameliorate TNBS-induced colitis and the PPARc ligand rosiglitazone has shown efficacy in mild to moderately active IBD. PPARc might also inhibit tumor growth in colorectal cancer. Alternatively, FABP1 has been suggested as a biomarker of CD because a 10fold up-regulation was found in one study. Our findings, in contrast, are consistent with loss of PPARc-mediated antiinflammatory effects. This supports a potentially important function for PPARc in the regulation of colonic inflammation and indicates that FABPs could also be a possible target for therapy. Among the 20 genes with highest FC at all time points were SPINK4; lipopolysaccharide binding protein, ADA and RETNLB. PCR validation was performed for SPINK4 and ADA. SPINK4 has recently been identified as a risk locus for UC and increased expression is seen in untreated celiac disease. LBP transfers LPS to the LPS-signaling receptor complex, promotes innate immunity against gram negative bacteria, and may serve as a marker of disease activity in CD. ADA 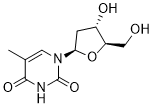 deactivates adenosine and thus diminishes the potentially protective effects of adenosine in mucosal inflammation and hypoxia. A beneficial effect after inhibition of ADA has been demonstrated in experimental colitis. RETNLB had a FC of 470 and 253 at T7 and T12, respectively.
deactivates adenosine and thus diminishes the potentially protective effects of adenosine in mucosal inflammation and hypoxia. A beneficial effect after inhibition of ADA has been demonstrated in experimental colitis. RETNLB had a FC of 470 and 253 at T7 and T12, respectively.