In EC by activating NFkB thus promoting pro-inflammatory 3,4,5-Trimethoxyphenylacetic acid angiogenesis, which has been associated with increased adventitial neovascularization and IH. IL-8 also leads to proliferation and migration 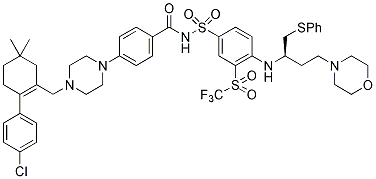 of vascular SMC thereby Chlorhexidine hydrochloride contributing to IH. IL-6 is a pro-inflammatory cytokine mainly secreted by activated macrophages and lymphocytes but also by EC and SMC. IL-6 is involved in immune regulation, hematopoiesis, inflammation and oncogenesis. Although little is known about the role of IL-6 in the pathophysiology of IH leading to vein bypass graft failure, several studies have demonstrated that IL-6 is pro-atherogenic through promoting EC dysfunction, SMC proliferation and migration as well as recruitment and activation of inflammatory cells. Besides IL-8 and IL-6 several other genes from these pathways have been associated with vascular remodeling and hence could affect vein graft implantation injury. In particular, we noted increased levels of transcriptional regulators such as NF-kB, cytokines and cytokine receptors such as IL1A, IL1B and IL1R2, regulators of extracellular matrix such as MMP2, COL1A1 and collagen type 1; and decreased levels of regulators of cell differentiation such as serum response factor. Based on the backpropagation networks, we also delineated the top focus gene hubs that had the greatest interaction density in both EC and SMC. The choice of these focus gene hubs was based on the fact that they provide maximum stability to the backpropagation network. In fact, targeting any of these genes as modeled by their removal from the network offers the most effective means to disrupt the network. Among those focus gene hubs, IL-6, INSR and IGF1R are the genes showing the greatest interaction density, closely followed by IL-8, IL-15 and FGFR2. In fact, among those genes, IL-6 and IL-8 were the most up-regulated genes, whereas INSR, IGF1R and FGFR2 were down-regulated. These results further validate the key role of IL-6 and IL-8 as pathogenic, and therefore as high profile therapeutic targets to prevent vein graft implantation injury. We have validated the up-regulation of IL-6 and IL-8 in graft EC and SMC by qRT-PCR, and confirmed that IL-6 significantly increases from 2 H and up to 7 D post-implantation while IL-8 is up-regulated at all time-points including the 30 D time-point in both cell types. Using qRT-PCR we also validated the upregulation of Coll11A1 at the later time-points, 7 and 30 D suggesting a potential role for extracellular remodeling in driving the healing process, while being the major component in lesion of implantation injury. Current work in our laboratory is aimed at developing local siRNA based therapies to concomitantly target IL-6 and IL-8 secretion within the vein graft and evaluate how this would impact vein graft implantation injury. In conclusion, this study represents the first comprehensive analysis of the genomic response to vein graft implantation injury in a large animal model. LCM has made it possible to separately define the genomic response of EC from that of medial SMC. Our data indicates that a robust genomic response begins by 2 H, peaks at 12�C24 H, starts resolving by 7 D, and declines markedly by 30 D. Inflammatory pathways dominate the early response, followed by modulators of cell cycling, and culminate in pathways involved in extra-cellular matrix remodeling. By using a back-propagation based systems biology analysis of the data, we were able to establish a temporal and causative link between these pathways that helped us identify the molecular signature of vein graft implantation injury, including high intensity hubs.
of vascular SMC thereby Chlorhexidine hydrochloride contributing to IH. IL-6 is a pro-inflammatory cytokine mainly secreted by activated macrophages and lymphocytes but also by EC and SMC. IL-6 is involved in immune regulation, hematopoiesis, inflammation and oncogenesis. Although little is known about the role of IL-6 in the pathophysiology of IH leading to vein bypass graft failure, several studies have demonstrated that IL-6 is pro-atherogenic through promoting EC dysfunction, SMC proliferation and migration as well as recruitment and activation of inflammatory cells. Besides IL-8 and IL-6 several other genes from these pathways have been associated with vascular remodeling and hence could affect vein graft implantation injury. In particular, we noted increased levels of transcriptional regulators such as NF-kB, cytokines and cytokine receptors such as IL1A, IL1B and IL1R2, regulators of extracellular matrix such as MMP2, COL1A1 and collagen type 1; and decreased levels of regulators of cell differentiation such as serum response factor. Based on the backpropagation networks, we also delineated the top focus gene hubs that had the greatest interaction density in both EC and SMC. The choice of these focus gene hubs was based on the fact that they provide maximum stability to the backpropagation network. In fact, targeting any of these genes as modeled by their removal from the network offers the most effective means to disrupt the network. Among those focus gene hubs, IL-6, INSR and IGF1R are the genes showing the greatest interaction density, closely followed by IL-8, IL-15 and FGFR2. In fact, among those genes, IL-6 and IL-8 were the most up-regulated genes, whereas INSR, IGF1R and FGFR2 were down-regulated. These results further validate the key role of IL-6 and IL-8 as pathogenic, and therefore as high profile therapeutic targets to prevent vein graft implantation injury. We have validated the up-regulation of IL-6 and IL-8 in graft EC and SMC by qRT-PCR, and confirmed that IL-6 significantly increases from 2 H and up to 7 D post-implantation while IL-8 is up-regulated at all time-points including the 30 D time-point in both cell types. Using qRT-PCR we also validated the upregulation of Coll11A1 at the later time-points, 7 and 30 D suggesting a potential role for extracellular remodeling in driving the healing process, while being the major component in lesion of implantation injury. Current work in our laboratory is aimed at developing local siRNA based therapies to concomitantly target IL-6 and IL-8 secretion within the vein graft and evaluate how this would impact vein graft implantation injury. In conclusion, this study represents the first comprehensive analysis of the genomic response to vein graft implantation injury in a large animal model. LCM has made it possible to separately define the genomic response of EC from that of medial SMC. Our data indicates that a robust genomic response begins by 2 H, peaks at 12�C24 H, starts resolving by 7 D, and declines markedly by 30 D. Inflammatory pathways dominate the early response, followed by modulators of cell cycling, and culminate in pathways involved in extra-cellular matrix remodeling. By using a back-propagation based systems biology analysis of the data, we were able to establish a temporal and causative link between these pathways that helped us identify the molecular signature of vein graft implantation injury, including high intensity hubs.
Monthly Archives: June 2019
Ephrin-A2 is expressed along the main cellular layers during embryonic the caudal tectum
Thus, one gradient of activity per axis is not sufficient to establish retinotopic LOUREIRIN-B connections, because a single cue gradient would cause all axons to migrate to and branch at one end of the map. Instead, counterbalanced forces are thought to be required, with each axon branching where these opposing forces balance out. Conflicting models have been postulated about the second mapping force. The most accepted model proposes that this second force is produced by a decreasing rostro-caudal gradient of EphA7 which repels nasal optic fibers and prevent them from branching in the rostral tectum/colliculus. However, as optic fibers invade the tectum/ colliculus throughout the highest part of this gradient, this model cannot explain how the axons invade the tectum/colliculus without being repelled by EphA7. Moreover, the role of tectal EphAs has not been evaluated in non mammalian vertebrates throughout in vivo experiments. Although differences in retinotectal/collicular mapping have been mainly described between fishes and amphibian versus birds and mammals, EphA3 and ephrin-A6 are only expressed in birds visual system and recent works suggest that some molecular mechanisms of retinotectal/ collicular mapping diverge between birds and mammals. Thus, the results obtained with collicular EphAs are not necessarily applicable to tectal EphA3. Given its decreasing rostro-caudal gradient in the chicken tectum, we hypothesized that EphA3 could account for the second activity necessary for retinotectal mapping along the rostro-caudal axis. Through functional in vitro and in vivo experiments, we demonstrated that the tectal gradient of EphA3 ectodomain is necessary to map nasal RGC axons on tectal surface by promoting nasal RGC axon growth toward the caudal Pimozide tectum and inhibiting branching rostrally to their appropriate termination zone. Furthermore, the promotion of axon growth by tectal EphA3 allows us to explain how the optic fibers invade the tectum. Therefore, opposite tectal gradients of EphA3 and ephrin-As counterbalance each other during retinotectal mapping. To investigate the function of tectal EphA3 we first analyzed its developmental pattern of expression by performing immunohistochemistry during the embryonic development 24 to 18 days of incubation – and the early postnatal period �Cnewly hatched, postnatal days 3 and 7 -. The study was achieved on sections coinciding with the rostro-caudal developmental gradient axis of the chicken optic tectum. This allowed us to distinguish between the most developed rostral pole, and the less developed caudal pole. We showed that EphA3 is expressed along the main cellular layers during the embryonic development. The stratum opticum formed by the optic fibers is also labeled mainly in the rostral tectum where the EphA3-positive temporal fibers arrive. The levels of expression of EphA3 in the 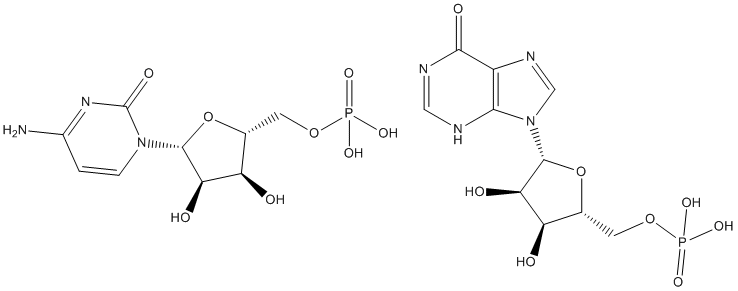 superficial layers �C which include the SO and the retino-recipient layers- produce a decreasing rostro-caudal gradient which extends along the entire tectal axis. This gradient can be appreciated at E5�CE6 �Cwhen the EphA3-positive temporal RGC axons have not yet invaded the tectum- and presents the highest level of expression around E12 when the retinotectal mapping is taking place. The EphA3 expression decreases postnatally when the retinotectal mapping has finished. As axonal ephrin-As are the potential receptors for tectal EphA3, we analyzed the developmental pattern of expression of ephrin-A2 by performing immunocytochemistry on retinal sections coinciding with the naso-temporal axis from E4 to P7.
superficial layers �C which include the SO and the retino-recipient layers- produce a decreasing rostro-caudal gradient which extends along the entire tectal axis. This gradient can be appreciated at E5�CE6 �Cwhen the EphA3-positive temporal RGC axons have not yet invaded the tectum- and presents the highest level of expression around E12 when the retinotectal mapping is taking place. The EphA3 expression decreases postnatally when the retinotectal mapping has finished. As axonal ephrin-As are the potential receptors for tectal EphA3, we analyzed the developmental pattern of expression of ephrin-A2 by performing immunocytochemistry on retinal sections coinciding with the naso-temporal axis from E4 to P7.
It is not easy to correlate the microarray data derived from planktonic cultures at least under our experimental conditions
It is possible that differences in chemotaxis might be appreciated by the use 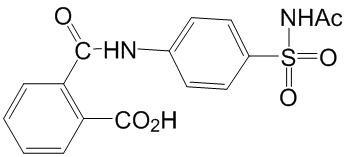 of specific attractant or repellent molecules. However, it is not trivial to identify such specific compounds and further studies should be performed in order to address this point. These unexpected and interesting results strongly suggest that the biological role of the RND-4 and RND-9 efflux pumps might not be restricted to the sole transport of toxic compounds, but also that their function might be Orbifloxacin related to motility and/or chemotaxis. To the best of our knowledge, this is the second time that the effect of RND efflux pumps mutation on motility-related phenotypes has been described. Indeed, the absence of RND components AcrB or TolC in Salmonella enterica caused widespread repression of chemotaxis and motility genes in these mutants, and for acrB mutant this was associated with decreased motility. However, why the deletion of an efflux pump should have a fallout on bacterial motility and chemotaxis remains an open question. It is conceivable that the cytoplasmic accumulation of efflux pump-specific metabolites could act as signals triggering opposite behavioural response in the two mutants. For instance, we have recently shown that RND-4 contributes to the transport of N-acyl homoserine lactone as we found a reduced accumulation of AHLs quorum sensing signal molecules in the growth medium of D4 mutant. Actually, the D4 and D4/D9 mutant produce about 30% less AHLs than the wild-type, while D9 produces almost the same level of acyl-HSL as the wild-type. In accordance with the low impact of D4 and D9 mutations on AHLs production, only few genes known to be AHLregulated are also differentially regulated in our microarray analysis. Among these, none can be directly related to chemotaxis or biofilm formation, and only BCAL0562 and BCAL3506 could be related to flagella. Overall, these observations suggest that it is unlikely that the phenotype of the D4, D9 and D4�CD9 mutants is due to an unbalance in AHLs import/export rates. However, it cannot be ruled out that other molecules acting as metabolic signals could accumulate in the D4, D9 and D4�CD9 mutants and account for the motility and biofilm phenotypes of these strains. Another possible explanation for the biological significance of the phenotype exhibited by D4 and D4�CD9 Gomisin-D strains might rely on the assumption that: i) the bacterial cell can ”sense” the concentration of toxic compounds outside and/or inside the cell and that ii) the cell itself tries to respond to the increase of the concentration of toxic compound by activating the efflux pump systems responsible for the extrusion of that compound. Accordingly, we can speculate that in the absence of these systems, the cell might somehow bypass this defect by increasing the ability to move in the environment in order to ”escape” and to explore spaces and niches where the concentration of the toxic compounds is lower. In other words, the increased ability to move might represent a sort of ”indirect protection” of the cell towards toxic compounds. Since in many bacteria flagellum could play a role in biofilm formation, the different regulation of flagellum-related genes in D4 and D9 prompted us to speculate that these strains might also have opposite biofilm phenotypes. Therefore, we performed preliminary experiments to investigate the biofilm formation ability of the wild-type and of the three mutants. Results showed, surprisingly, that all the mutants had an enhancement of biofilm formation with respect to the wild-type. Therefore, differences in flagella expression in the D4 and D9 strains, with respect to the wildtype, play a minor role in biofilm formation, at least under our experimental conditions. The increased biofilm production of the RND-mutants was unexpected since we did not identify genes obviously involved in biofilm formation among the 33 having the same expression pattern in the three microarray experiments. Actually, biofilm formation is a complex pleiotropic phenotype, strongly dependent upon experimental conditions and growth media.
of specific attractant or repellent molecules. However, it is not trivial to identify such specific compounds and further studies should be performed in order to address this point. These unexpected and interesting results strongly suggest that the biological role of the RND-4 and RND-9 efflux pumps might not be restricted to the sole transport of toxic compounds, but also that their function might be Orbifloxacin related to motility and/or chemotaxis. To the best of our knowledge, this is the second time that the effect of RND efflux pumps mutation on motility-related phenotypes has been described. Indeed, the absence of RND components AcrB or TolC in Salmonella enterica caused widespread repression of chemotaxis and motility genes in these mutants, and for acrB mutant this was associated with decreased motility. However, why the deletion of an efflux pump should have a fallout on bacterial motility and chemotaxis remains an open question. It is conceivable that the cytoplasmic accumulation of efflux pump-specific metabolites could act as signals triggering opposite behavioural response in the two mutants. For instance, we have recently shown that RND-4 contributes to the transport of N-acyl homoserine lactone as we found a reduced accumulation of AHLs quorum sensing signal molecules in the growth medium of D4 mutant. Actually, the D4 and D4/D9 mutant produce about 30% less AHLs than the wild-type, while D9 produces almost the same level of acyl-HSL as the wild-type. In accordance with the low impact of D4 and D9 mutations on AHLs production, only few genes known to be AHLregulated are also differentially regulated in our microarray analysis. Among these, none can be directly related to chemotaxis or biofilm formation, and only BCAL0562 and BCAL3506 could be related to flagella. Overall, these observations suggest that it is unlikely that the phenotype of the D4, D9 and D4�CD9 mutants is due to an unbalance in AHLs import/export rates. However, it cannot be ruled out that other molecules acting as metabolic signals could accumulate in the D4, D9 and D4�CD9 mutants and account for the motility and biofilm phenotypes of these strains. Another possible explanation for the biological significance of the phenotype exhibited by D4 and D4�CD9 Gomisin-D strains might rely on the assumption that: i) the bacterial cell can ”sense” the concentration of toxic compounds outside and/or inside the cell and that ii) the cell itself tries to respond to the increase of the concentration of toxic compound by activating the efflux pump systems responsible for the extrusion of that compound. Accordingly, we can speculate that in the absence of these systems, the cell might somehow bypass this defect by increasing the ability to move in the environment in order to ”escape” and to explore spaces and niches where the concentration of the toxic compounds is lower. In other words, the increased ability to move might represent a sort of ”indirect protection” of the cell towards toxic compounds. Since in many bacteria flagellum could play a role in biofilm formation, the different regulation of flagellum-related genes in D4 and D9 prompted us to speculate that these strains might also have opposite biofilm phenotypes. Therefore, we performed preliminary experiments to investigate the biofilm formation ability of the wild-type and of the three mutants. Results showed, surprisingly, that all the mutants had an enhancement of biofilm formation with respect to the wild-type. Therefore, differences in flagella expression in the D4 and D9 strains, with respect to the wildtype, play a minor role in biofilm formation, at least under our experimental conditions. The increased biofilm production of the RND-mutants was unexpected since we did not identify genes obviously involved in biofilm formation among the 33 having the same expression pattern in the three microarray experiments. Actually, biofilm formation is a complex pleiotropic phenotype, strongly dependent upon experimental conditions and growth media.
It is plausible that various stem cells employ common mechanisms through the use of evolutionarily related families of proteins
The inclusion of evolutionarily related genes as modules revealed a striking number of families with expression patterns associated with stemness. Duplication and subsequent modification of genes and their enhancers through evolution may have provided the control logic needed to diversify stem cells for populating new tissue systems distinct from ancestral counterparts. Indeed, the homolog modules consistently received higher S-MAP scores compared to the modules in a previous study that did not consider gene homology. The identification of stemness modules has obvious implications for both iPS cells and cancer therapeutics. Reprogramming candidates essential for stem cell properties are likely found among the stemness-on modules while stemness-off modules likely contain genes that have to be silent in stem cells. The SI scoring strategy, possibly adapted specifically to ESC/iPS cell signatures, could be used in the characterization of putative iPS lines. Similar strategies can be used to identify and target CSC within a heterogeneous cancer population and serve as valuable tools for fighting cancer progression. In addition, the results can be used to test for novel stem cell marker genes to improve the definition and isolation of stem cells. The S-MAP approach synergizes with large-scale efforts to systematically extract differentially expressed genes in many different conditions represented in current microarray gene expression repositories, such as Oncomine and GeneChaser. It is complementary to previous approaches to uncover the mechanisms of pluri- and multi-potency maintenance, such as the development of the PluriNet. In this study, we focused our analysis on modules with expression patterns ranging across a broad array of different stem cell types. However, S-MAP also revealed 152 homolog and 32 functional modules that were restricted to specific stem cell types; these modules should reveal tissue- or lineage-specific stem cell mechanisms. The method synergizes with large-scale efforts to systematically extract differentially expressed genes in many different conditions represented in current microarray gene expression repositories, such as Oncomine and GeneChaser. Finally, S-MAP can be used to test for inter-species stem cell expression Ginsenoside-F2 conservation, to characterize cancers from their normal counterparts, and is readily transferrable to other systems and diverse types of data. While the S-MAP procedure could score all collected modules, we defined a nonredundant set of modules to use for assessing the overall significance and distribution of scores. Allowing Albaspidin-AA redundancy could skew the results because of highly characterized pathways. For example, Gene Ontology contains a deep hierarchy with many sub- and sub-sub-categorizations under the cell cycle process because it has been studied extensively in yeast. A redundancy filter on the functional modules helped us avoid redundant annotations of highly characterized pathways and protein complexes from dominating the results. To filter out redundant functional modules, we first sorted the functional modules from smallest to largest and excluded any having a 25% or greater gene overlap with a smaller functional module. Functional modules with over half of the genes belonging to the same set of homologs were also excluded because any such module also reflects an evolutionary connection between the genes to a greater degree than the functional connection. The relatively high overlap cutoff between evolutionary and functional gene modules allows genes with multiple functional roles to be captured by different module types. The recurrence score used for S-MAP was motivated by the need to compensate for potential biases in combining heterogeneous results. There are obvious variations in the overlap and sizes of the SGLs and DGLs, which reflect the differences in the compared 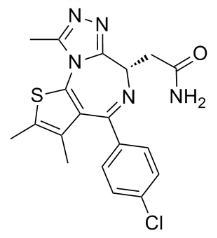 biological specimens, cell, and RNA isolation techniques, hybridization protocols, and statistical methods applied to identify differentially expressed clones.
biological specimens, cell, and RNA isolation techniques, hybridization protocols, and statistical methods applied to identify differentially expressed clones.
The influence of studies reporting highly similar gene lists to avoid a bias due to over-representation of any one stem cell type
Before computing recurrence scores for genes or modules, we calculated the similarity of all gene lists to derive groups of gene lists. Gene lists in the same group could then be collectively down-weighted in the recurrence scoring. The identification of these gene list groups is described next. DNA is a dynamic molecule and is constantly exposed to various types of damaging agents such as mutagenic chemicals, radiation and reactive oxygen. A number of DNA repair systems exist which specialize in the repair of certain types of damage. Nucleotide excision repair is a highly conserved pathway involved in repair of a wide variety of structurally unrelated DNA lesions. One of the well characterized NER systems is the UvrABC nuclease from E. coli. NER consists of two related sub-pathways; global genomic repair, which removes lesions from the overall genome, and transcription coupled repair, which removes lesions from the transcribed strand of active genes. Bulky DNA lesions such as cyclo pyrimidine photodimers induced by UV irradiation block RNA polymerase during transcription. In bacteria a product of mfd called transcription repair coupling factor or Mfd protein is required for TCR. Bacterial Mfd interacts with the stalled RNA polymerase, displaces it from the DNA and recruits NER proteins at the site of damage. Mfd thus clears the steric hindrance from the site of damage and loads UvrA protein, resulting in,10-fold faster repair of the transcribed strand compared to the non-transcribed strand for similar kind of lesions. In addition, Mfd rescues arrested or backtracked transcription elongation complexes by promoting forward translocation of RNA polymerase in ATP dependent manner leading to productive elongation. Additionally, Mfd can release the RNA polymerase when the enzyme cannot continue elongation. Apart from DNA repair, Mfd has other physiological roles in regulation of gene expression, including carbon catabolite repression in Bacillus subtilis  and transcription termination by bacteriophage HK022 Nun protein. A key role for Mfd as an enhancer of UvrA turnover in E. coli cells has also been recently demonstrated. The well characterized Mfd from E. coli is a 130 kDa monomeric protein having modular architecture specialized for different functions. The N-terminal domain shares a high degree of structural homology with UvrB protein of NER pathway. The NTD is known to interact with UvrA protein, which is molecular matchmaker of NER pathway, and this interaction is responsible for enhanced rates of repair. The central portion of Mfd consists of RNA polymerase interacting domain which binds to b subunit of RNA polymerase. The C-terminal domain of Mfd harbors seven signature motifs of super-family 2 helicases including ATPase motifs. In addition, CTD contains a TRG motif required for translocation along the DNA. TRG motif as the name implies, is highly homologous to RecG protein, which is known to be involved in branch migration of Holliday junctions during recombination. Pathogenic bacteria continuously encounter multiple forms of stress in their hostile environments, which leads to DNA damage. Genes involved in DNA repair and recombination may play an important role in the virulence of pathogenic organisms. M. tuberculosis is a gram positive, acid fast bacterium and one of the most formidable human pathogen. DNA repair pathways in mycobacteria appear to be crucial for their survival at different stages of infection. Sequencing of M. tuberculosis genome revealed the presence of NER associated genes including a putative mfd. In this work, we describe the functional characterization of MtbMfd and discuss its unusual properties. This is the first detailed analysis of the biochemical Ginsenoside-F4 properties of Mfd from actinomycetes and more importantly from a human pathogen. Limited proteolysis is often employed to determine the domainal organization, stability and Folinic acid calcium salt pentahydrate conformational changes within the protein. The hexamer and monomer fractions of MtbMfd obtained by gel filtration chromatography were subjected to limited digestion by trypsin.
and transcription termination by bacteriophage HK022 Nun protein. A key role for Mfd as an enhancer of UvrA turnover in E. coli cells has also been recently demonstrated. The well characterized Mfd from E. coli is a 130 kDa monomeric protein having modular architecture specialized for different functions. The N-terminal domain shares a high degree of structural homology with UvrB protein of NER pathway. The NTD is known to interact with UvrA protein, which is molecular matchmaker of NER pathway, and this interaction is responsible for enhanced rates of repair. The central portion of Mfd consists of RNA polymerase interacting domain which binds to b subunit of RNA polymerase. The C-terminal domain of Mfd harbors seven signature motifs of super-family 2 helicases including ATPase motifs. In addition, CTD contains a TRG motif required for translocation along the DNA. TRG motif as the name implies, is highly homologous to RecG protein, which is known to be involved in branch migration of Holliday junctions during recombination. Pathogenic bacteria continuously encounter multiple forms of stress in their hostile environments, which leads to DNA damage. Genes involved in DNA repair and recombination may play an important role in the virulence of pathogenic organisms. M. tuberculosis is a gram positive, acid fast bacterium and one of the most formidable human pathogen. DNA repair pathways in mycobacteria appear to be crucial for their survival at different stages of infection. Sequencing of M. tuberculosis genome revealed the presence of NER associated genes including a putative mfd. In this work, we describe the functional characterization of MtbMfd and discuss its unusual properties. This is the first detailed analysis of the biochemical Ginsenoside-F4 properties of Mfd from actinomycetes and more importantly from a human pathogen. Limited proteolysis is often employed to determine the domainal organization, stability and Folinic acid calcium salt pentahydrate conformational changes within the protein. The hexamer and monomer fractions of MtbMfd obtained by gel filtration chromatography were subjected to limited digestion by trypsin.