Thus, like Insv, Insb appears to SJN 2511 function in a partly redundant manner with H. Additionally, while loss of insb and insv activities similarly enhanced the H haplo-insufficient Masitinib VEGFR/PDGFR inhibitor phenotype, no genetic interaction was observed in double mutant flies. One possible interpretation for this lack of genetic interaction is that Insv and Insb act together to regulate the same process, so that the complete loss of one or both genes have similar 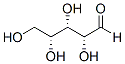 phenotypic consequences. Since Insv did not regulate the expression of insb, one possibility is that Insb positively regulates the expression of the insv gene and that Insv antagonizes Notch. Alternatively, the two proteins may act together to repress the expression of Notch target genes via the Su binding sites. Consistent with this, Insv was proposed to repress the expression of Notch target genes by two mechanisms: first in a Su-dependent mechanims, Insv would act as a CSL co-repressor to promote repression through Su binding sites; second, Insv may directly bind DNA via its BEN domain and regulate gene expression in a Su-independent manner. Whether Insb physically interacts with Insv and regulates its transcriptional activities await biochemical studies. While a functional homolog of Insv has recently been characterized in the mouse, no clear homolog of Insb could be easily identified in vertebrates. Thus, deciphering how Insb regulates in flies the activities of Insv and other CSL associated co-repressors, such as H, may provide new insights into molecular mechanisms of co-repression by CSL-associated factors. Finally, while the expression and function of Insb was primarily studied here in the context of sensory organ development, this gene was also expressed at high levels in neuroblasts of the developing larval brain, suggesting that Insb may have a broader role as a Notch antagonist. In conclusion, our study identified Insb as a nuclear SOP/ neuron-specific antagonist of Notch signaling that may act together with Insv to repress the expression of Notch target genes. Assembly of the highly conserved tubulin-like protein FtsZ into a ring structure at the nascent division site initiates the process of cell division in most bacteria. The FtsZ ring serves as a foundation for assembly of the division machinery and constricts at the leading edge of the invaginating septum during cytokinesis. The precise temporal and spatial regulation of cell division is achieved through the actions of a host of proteins, which interact directly with FtsZ to modulate assembly of the cytokinetic ring. Some of these modulators help stabilize FtsZ polymers at midcell and thus maintain the integrity of the cytokinetic ring. In both Bacillus subtilis and Escherichia coli, the location of FtsZ ring formation appears to be dictated in part through the actions of proteins that inhibit FtsZ assembly at aberrant subcellular positions. In B. subtilis, EzrA, a 65 kDa membrane bound protein, plays an important role in both modulatory roles. EzrA is among the first set of proteins to localize to the cytokinetic ring. Null mutations in ezrA reduce the critical concentration of FtsZ required for ring formation in vivo and result in the formation of extra FtsZ rings and septa at cell poles. In contrast to loss of function mutations in other positional regulators of bacterial cell division, the loss of EzrA significantly increases the stability of the medial FtsZ ring, rendering it resistant to overexpression of division inhibitors. Null mutations in ezrA or a point mutation that disrupts EzrA localization to midcell increase cell length by more than 50%, consistent with a model in which EzrA is required for the efficient use of the medial division site.
phenotypic consequences. Since Insv did not regulate the expression of insb, one possibility is that Insb positively regulates the expression of the insv gene and that Insv antagonizes Notch. Alternatively, the two proteins may act together to repress the expression of Notch target genes via the Su binding sites. Consistent with this, Insv was proposed to repress the expression of Notch target genes by two mechanisms: first in a Su-dependent mechanims, Insv would act as a CSL co-repressor to promote repression through Su binding sites; second, Insv may directly bind DNA via its BEN domain and regulate gene expression in a Su-independent manner. Whether Insb physically interacts with Insv and regulates its transcriptional activities await biochemical studies. While a functional homolog of Insv has recently been characterized in the mouse, no clear homolog of Insb could be easily identified in vertebrates. Thus, deciphering how Insb regulates in flies the activities of Insv and other CSL associated co-repressors, such as H, may provide new insights into molecular mechanisms of co-repression by CSL-associated factors. Finally, while the expression and function of Insb was primarily studied here in the context of sensory organ development, this gene was also expressed at high levels in neuroblasts of the developing larval brain, suggesting that Insb may have a broader role as a Notch antagonist. In conclusion, our study identified Insb as a nuclear SOP/ neuron-specific antagonist of Notch signaling that may act together with Insv to repress the expression of Notch target genes. Assembly of the highly conserved tubulin-like protein FtsZ into a ring structure at the nascent division site initiates the process of cell division in most bacteria. The FtsZ ring serves as a foundation for assembly of the division machinery and constricts at the leading edge of the invaginating septum during cytokinesis. The precise temporal and spatial regulation of cell division is achieved through the actions of a host of proteins, which interact directly with FtsZ to modulate assembly of the cytokinetic ring. Some of these modulators help stabilize FtsZ polymers at midcell and thus maintain the integrity of the cytokinetic ring. In both Bacillus subtilis and Escherichia coli, the location of FtsZ ring formation appears to be dictated in part through the actions of proteins that inhibit FtsZ assembly at aberrant subcellular positions. In B. subtilis, EzrA, a 65 kDa membrane bound protein, plays an important role in both modulatory roles. EzrA is among the first set of proteins to localize to the cytokinetic ring. Null mutations in ezrA reduce the critical concentration of FtsZ required for ring formation in vivo and result in the formation of extra FtsZ rings and septa at cell poles. In contrast to loss of function mutations in other positional regulators of bacterial cell division, the loss of EzrA significantly increases the stability of the medial FtsZ ring, rendering it resistant to overexpression of division inhibitors. Null mutations in ezrA or a point mutation that disrupts EzrA localization to midcell increase cell length by more than 50%, consistent with a model in which EzrA is required for the efficient use of the medial division site.
Biochemical experiments indicate that EzrA interacts directly with FtsZ to inhibit assembly
Leave a reply