Adding biomarker testing as a secondary endpoint to an ongoing phase study represented a timely and . In particular, evidence for a biomarker typically does not KRX-0401 appear early in the drug development process; instead, it usually emerges during phase 2 evaluation and often after a phase 3 study has been initiated. In our case, the PLGF biomarker hypothesis was developed in early-phase testing, with analysis of the phase 2 data occurring while a phase 3 study was ongoing. Consequently, the PLGF hypothesis was added to the phase 3 study following interactions with the FDA. While the option of assessing PLGF as a predictive pharmacodynamic biomarker for motesanib in a larger, independent phase 2 study first represented a scientifically ideal approach, it would have resulted in significant delays in evaluating the hypothesis with no guarantee of a positive outcome. Potentially, a confirmatory prospective run-in design trial may have been considered had the PLGF biomarker hypothesis been confirmed in MONET1. It has been suggested that less than 1% of published cancer biomarkers are routinely used in the clinical setting. Factors identified as contributing to failure to translate biomarkers into the clinic include lack of clinical practicality of the biomarker, hidden biases within the data, an inadequate assay, inappropriate statistical techniques, and lack of biologic plausibility for the biomarker. Although we were not successful in developing a predictive biomarker for motesanib in NSCLC our approach adequately addressed these factors. Biomarker identification was included in early-phase studies, we developed adequate statistical techniques, a robust diagnostic test 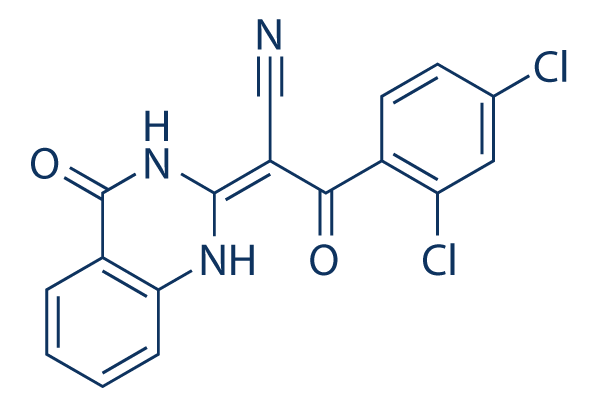 to evaluate PLGF, and engaged early with the US FDA to gain support for our protocol amendment. However, using a pharmacodynamic biomarker as a predictor of efficacy remains an unproven approach. Such biomarkers have typically only been used to identify toxicity issues and there is no precedent that could have guided the development of the biomarker portion of our study. Our experience illustrates several significant challenges to develop predictive pharmacodynamic biomarkers in oncology. Ideal approaches calling for specific study designs and/or sequences of events should be applied wherever possible in an effort to maximize the chances of PB 203580 p38 MAPK inhibitor success; however, they seldom reflect the unpredictable scenarios that may unfold during drug development. Furthermore, a methodical, no-risk approach must be balanced against economic factors and the desire to rapidly identify patient populations that may benefit the most from a potential new treatment. Despite these challenges, it remains important to develop biomarker hypotheses and to subject them to objective evaluation in clinical studies. Development of predictive pharmacodynamic biomarkers remains an opportunity to markedly improve outcomes for patients. With more than 400 millions infections worldwide, malaria remains a major public health issue, principally in sub-Saharan Africa. An effective vaccine would help reduce disease burden, but the best candidates are still in development or evaluation phase. The rapid development of multidrug-resistant Plasmodium parasites necessitates accelerating the discovery of novel antimalarial compounds to meet the needs of the agenda for malaria control and eradication. In humans, Plasmodium sp. development comprises different stages, with the asexual intra�Cerythrocytic forms being responsible for the symptoms of the disease, such as fever, anemia, and cerebral malaria that can lead to death. The erythrocyte invasion by Plasmodium merozoites critically depends on protease activities involved in both the daughter parasites egress from erythrocytes, and invasion into another erythrocyte.
to evaluate PLGF, and engaged early with the US FDA to gain support for our protocol amendment. However, using a pharmacodynamic biomarker as a predictor of efficacy remains an unproven approach. Such biomarkers have typically only been used to identify toxicity issues and there is no precedent that could have guided the development of the biomarker portion of our study. Our experience illustrates several significant challenges to develop predictive pharmacodynamic biomarkers in oncology. Ideal approaches calling for specific study designs and/or sequences of events should be applied wherever possible in an effort to maximize the chances of PB 203580 p38 MAPK inhibitor success; however, they seldom reflect the unpredictable scenarios that may unfold during drug development. Furthermore, a methodical, no-risk approach must be balanced against economic factors and the desire to rapidly identify patient populations that may benefit the most from a potential new treatment. Despite these challenges, it remains important to develop biomarker hypotheses and to subject them to objective evaluation in clinical studies. Development of predictive pharmacodynamic biomarkers remains an opportunity to markedly improve outcomes for patients. With more than 400 millions infections worldwide, malaria remains a major public health issue, principally in sub-Saharan Africa. An effective vaccine would help reduce disease burden, but the best candidates are still in development or evaluation phase. The rapid development of multidrug-resistant Plasmodium parasites necessitates accelerating the discovery of novel antimalarial compounds to meet the needs of the agenda for malaria control and eradication. In humans, Plasmodium sp. development comprises different stages, with the asexual intra�Cerythrocytic forms being responsible for the symptoms of the disease, such as fever, anemia, and cerebral malaria that can lead to death. The erythrocyte invasion by Plasmodium merozoites critically depends on protease activities involved in both the daughter parasites egress from erythrocytes, and invasion into another erythrocyte.
Monthly Archives: July 2019
Ran conformational sampling of the mutant with molecular dynamics and calculated the free energy of binding
In this step, we mutated residues in EETI-II-sub at the protein�Cprotein interface of the complex via implicit solvent models based on the Generalized Born approximation. The last step consisted in the experimental testing of the inhibitor by an enzymatic inhibitory assay specific for the PvSUB1 recombinant enzyme. A preliminary free energy calculation was performed with snapshots from multiple MD simulations of EETI-II-sub docked onto PvSUB1 to obtain more Ruxolitinib consistent MM/GBSA results. A free-energy decomposition shows the contribution of each single residue to the total free energy of binding. The biggest contribution to the free energy of binding came from the main-chain contacts of residues P4, P3, P2 and P1. This is in agreement with previous observations of important interactions between a protein-inhibitor and a serine protease active site, where important contacts are made by main-chain atoms. For the case of EETI-II-sub the highest contribution originates from the cysteine in P3 and its main-chain, accounting for 24.34 kcal/ mol. We then tried to identify the most favorable SCH772984 ERK inhibitor mutations that could improve the binding affinity of EETI- II-sub to PvSUB1. The cysteine in P3 cannot be mutated because its side-chain is involved in a disulphide bridge that has an important function in stabilizing the EETI-II scaffold and maintaining the loop rigid, whereas the alanine in P2 already contributes with 24.17 kcal/ mol to the total binding energy. We also looked at the parasite sequences that are natural substrates of PvSUB1 or PfSUB1 to suggest positions to introduce mutations in the EETI-II scaffold. Table 4 lists these sequences for PfSUB1 and PvSUB1. While the sequences of several PfSUB1 substrates were experimentally determined, few were identified for PvSUB1. Considering the evolutionary proximity of P. vivax and P. falciparum, with active sites displaying.60% sequence identity, these predicted sequences can be considered reliable. Comparing the cleavage sites, we observed that only alanine and glycine appeared in P2, suggesting that only small residues are tolerated in this position. Position P19 has a negative contribution to the energy and therefore is an interesting position to mutate. However, considering the lack of a specific pocket for this residue we can consider this position almost a secondary contact residue and we decided to keep the wild�Ctype residue. Finally, SUB1 cleavage site sequences have a fairly high similarity at the P1 and P4 positions and we therefore focused on these positions to mutate the EETI-II inhibitory loop. It is worth mentioning that the contribution of these P1 and P4 positions within the substrate-PfSUB1 interaction has recently been experimentally established. We performed 106100 ps MD  simulations and MM/GBSA free energy calculations for all possible residues in position P4 and P1 independently, assuming that the effect of the two mutations would be additive. The free energy calculations for the mutants in P4 showed that hydrophobic and bulky residues were preferred for this position. This result fits with the fact that pocket S4 is composed of six hydrophobic residues and seems to have enough space to accommodate larger hydrophobic side-chains than valine. Position P1 instead presents as favorable mutations aromatic residues with polar groups, glutamate and positively charged residues. Surprisingly we found as favorable mutations some positively charged residues, whereas most of sequences recognized by the homologous PfSUB1 present negatively charged or neutral polar sidechains at P1. This might be explained by either the low substrate specificity common to some subtilisins or imprecisions in the structure of the complex.
simulations and MM/GBSA free energy calculations for all possible residues in position P4 and P1 independently, assuming that the effect of the two mutations would be additive. The free energy calculations for the mutants in P4 showed that hydrophobic and bulky residues were preferred for this position. This result fits with the fact that pocket S4 is composed of six hydrophobic residues and seems to have enough space to accommodate larger hydrophobic side-chains than valine. Position P1 instead presents as favorable mutations aromatic residues with polar groups, glutamate and positively charged residues. Surprisingly we found as favorable mutations some positively charged residues, whereas most of sequences recognized by the homologous PfSUB1 present negatively charged or neutral polar sidechains at P1. This might be explained by either the low substrate specificity common to some subtilisins or imprecisions in the structure of the complex.
We determined average peak amplitudes of the GABA response of oocytes treated with a given scFv
In the first of these comparisons, and normalized these values to the average peak amplitudes obtained from control GABAA expressing oocytes. Entries in columns 1 and 2 of Table 2 show the normalized peak amplitude of the GABA-elicited response for a given treatment. Entries in columns 3 and 4 indicate normalized peak amplitudes of the GABA-elicited response for each scFv after incubation with secondary antibody. This was done to study the effect on channel dynamics of adding a macromolecular, sterically bulky moiety to the scFv. The results indicate that the membrane current responses of oocytes incubated with and without the secondary antibody reagent are not substantially different. The second measure of comparison performed was to analyze the effects of a given treatment on the waveform of the GABA-elicited response. Determinations of L provide further information on the effect of scFv treatment. They show, specifically, that point-by-point comparison of peak-matched waveforms of untreated and treated oocytes yield goodness-of-fit values near unity, indicating high similarity in waveform kinetics, consistent with minimal or no effect on receptor channel activity upon treatment with scFvs. While physiological control of the receptor by a combinatorially-derived affinity reagent has been observed in previous studies, the absence of an effect of the scFvs studied here on the GABAA electrophysiological activity is consistent with the suitability of these scFvs as the anchor component of a prospective neuromodulator. In immunohistochemistry experiments on cryosections of mouse retina, we investigated the binding of A10, the a1-specific scFv described above. The results,U0126 shown in Figure 4A, indicate distinct staining of the ganglion cell and inner plexiform layers, consistent with previously reported data. A similar staining pattern was obtained with commercially available anti-a1 monoclonal antibody. The small differences in staining patterns obtained with the present scFv and the commercial antibody can be explained by possible differences in the epitopes used to generate the two antibodies. Further evidence that the staining by A10 is due to specific recognition of the receptor came from an experiment in which we tested an scFv raised against an unrelated target ]. This ZF130H1-directed scFv did not yield a layer-specific staining pattern in retina. These data demonstrate that the scFv A10 recognizes GABAA a1 subunit in the mouse retina. The present study demonstrates the successful isolation of scFvs against GABAA receptor subunits by phage display technology using synthetic peptides. These scFvs bind to their cognate peptide specifically, with minimal or no cross-reactivity to peptides of other GABA subunits. Significantly, these peptide-raised scFvs specifically bind intact GABAA receptors expressed in Xenopus oocytes, and appear not to substantially alter the receptor’s electrophysiological activity. Furthermore, A10, the a1-specific scFv,VE-822 labels mouse retina in layers expected to contain GABAA receptors. The results additionally show that dimerization of the A10 scFv yields a more robust reagent, as evidenced by the10-fold enhancement of sensitivity in dot blots and the more than 100-fold reduction in Kd value.
Since our DiME method only extracts a single best module by definition connected subgraph of the entire graph
The estimation and seeding process used in our algorithm is relatively simple and straightforward, we could for each extraction procedure view each node as a possible ‘‘seed’’ for the module to be extracted, which will progressively include its surrounding nodes to form the module during optimisation. The initial extraction with relatively few individuals would, then, act as the seed prioritizer. To ensure that the modules extracted from the biological networks are statistically significant, i.e. they are significantly different from modules that arise from random networks of an appropriate null model, we incorporated a B-score significance measure as proposed in as a quality control step for the modules. The B-score measure assumes a null model where edges within the module of PI-103 interest is held unchanged while the remaining connections in the network are randomly shuffled. Then the B-score is calculated based on the null module to quantify how often we should expect to see the module ‘‘by chance’’. The B-scoring measure has a major advantage of avoiding large amounts of resampling cycles for simulating null model results. In our later experiments, we also showed that the Bscoring measure worked well with our DiME algorithm to detect statistically significant modules. For details of the B-score calculation, the reader is referred to the original works. In order to make this paper selfcontained, we provide the full procedure for B-score computation in Section S1 in File S1. In this study all B-score calculations were based upon default parameters in the original work with 20 independent runs for each module evaluation. We then selected the a certain percentage of the top ranking gene pairs as significant co-expressions, which will be connected as a coexpression network. This percentage, called network construction threshold in our paper, will affect the edge noise level of the resulting network. For example, PLX4032 a stringent threshold will miss large numbers of true edges while a larger threshold will introduce many false-positive edges. For our glioma co-expression network analysis, we set the network construction threshold to 0.1%. In the Results section, we also present data regarding how different network construction threshold values, therefore different noise levels, affect module extraction results of the DiME algorithm. Edge widths are designed to be proportional to the number of connections between two modules, in order to illustrate strength of coordination between functional components in the disease network. Node color represents fold change of average expression level of all genes in one module compared with normal patient samples. The grade II glioma module network demonstrates a significant shift in the tumour phenotype compared with normal samples. As would be expected, there appears to be a marked down-regulation of normal neuronal function, and a significant increase in cell cycle-associated processes. It is of interest to note that the modules associated with immune response are slightly, but significantly increased in grade II tumours.
They reported that DD genotype had higher risk of CKD than ID genotype, followed by the II genotype
Studies of Caucasian subjects have indicated additive effects of the D allele in females, but studies of Asian subjects have shown different results. Although many previous metaanalysis studies investigating ACE I/D polymorphisms and CKD have been reported, but no studies have considered moderate effects of gender in our knowledge. This study focused on general population without genetic abnormality or rare disorder, and we wanted to compare the risk of CKD in people with major allele or minor allele on ACE I/D. In addition, gender-dependent effects of ACE I/D polymorphisms on CKD risk was investigated. This study showed that CKD risk was higher in D allele carriers than in I allele carriers, and there was no strong evidence that analyses using different model assumptions might produce dissimilar results. Heterogeneity was higher in the Asian population than in the Caucasian population. Interaction between ACE I/D polymorphisms and hypertension exerted an additive effect on CKD risk. A gender-dependent effect of ACE I/D polymorphisms on CKD risk was clearly apparent in Asians but not in Caucasians. The DD genotype showed higher gene expression and serum ACE levels than the ID genotype, followed by the II genotype. High blood ACE levels may increase blood angiotensin II levels, and individuals with higher angiotensin II levels may have a higher CKD risk. Previous studies showed that the association between ACE I/D polymorphisms and CKD risk might not be dominant or LY2157299 recessive. Previous metaanalysis studies showed the supported results. We also observed the apparent linear association between numbers of D allele and odds ratios compared the II genotype in genotype analyses. The assumption of the allele type model in this association might be more reasonable, and it may thus be true that individuals carrying the D allele have a higher CKD risk. Hypertension in some patients is due to a dysfunction of RAS such as abnormal secretion of renin, causing increased blood angiotensin I levels. D allele carriers had higher ACE levels than I allele carriers, leading to more efficient conversion of angiotensin I to angiotensin II,LY294002 resulting in CKD. The mechanism may be an additive effect of hypertension and the D allele. An additive effect was significant in the nondiabetic group but not in the diabetic nephropathy subgroups. The blood levels of advanced glycation end products diabetic patients may be high, possibly causing blood pressure increases. We accordingly hypothesize that the probability of hypertension because of a dysfunction of RAS was higher in the nondiabetic nephropathy subgroup than in the diabetic nephropathy subgroup. Thus, the interaction between ACE I/D polymorphisms and hypertension was significant only in the nondiabetic nephropathy subgroup. This hypothesis may require further studies for confirmation. We found a significant gender-dependent effect of ACE I/D polymorphisms on CKD risk in Asians. In previous studies in Asians, the ORs of the additive effect on the DD genotype of males were 2.94 and 1.41 in Japanese and Koreans, respectively. Another study in Japan also reported a positive additive effect of the DD genotype of males. Studies of Caucasians reported contrary results, with an interaction OR of 0.42 in Pakistan. Another two studies in France and Mexico also showed an additive effect between the DD genotype and female gender but not male gender.